



What is Python?
As a general-purpose programming language, Python is universal. It’s quick and easy, yet powerful with plenty of capabilities. It gives you an opportunity to build your machine learning/deep learning models, web applications, and anything else you need in one language.
Scientific Computing Libraries
Owing to its ease-of-use, versatility, and the availability of mature scientific computing libraries like NumPy, SciPy, etc; Python has become the standard language for Data Science. Python also offers a wide range of free machine learning libraries like SciKit-learn and data analysis libraries like Pandas. Evidently, Pandas DataFrame abstraction has provided the motivation and inspiration to develop Apache Spark Dataframe abstraction tailored for in-memory cluster computing. A similar pattern can be found with another component of Spark; Spark MLlib library that draws inspiration from SciKit-learn. Spark MLlib later evolved to Spark ML pivoting from the legacy RDD abstraction to a DataFrame abstraction.
Cluster Computing
Over 90% of all the data in the world has been created in just the last 2 years. This unprecedented growth in volumes of data is pushing Data Analysts and Data Scientists working in Python to extend and scale the single-node computation of Python to distributed cluster computational frameworks like Apache Spark.
In this blog, we will cover some recipes to extend frameworks like scikit-learn and provide parallelism over a distributed spark cluster. We will also explore additional recipes to boost parallelism using the Python Multiprocessing library with PySpark SparkML library.
Scikit-Learn
Scikit-learn uses joblib for single-machine parallelism. This lets you train most estimators (anything that accepts the n_jobs parameter) using all the cores of your laptop or workstation.
Joblib has an Apache Spark extension: joblib-spark. Scikit-learn can use this extension to train estimators in parallel on all the workers of your spark cluster without significantly changing your code. Note that, this requires scikit-learn>=0.21 and pyspark>=2.4
Training the estimators using Spark as a parallel backend for SciKit-Learn is most useful in the following scenarios.
- Training a Large Model on Medium-Sized datasets that can fit in the memory of a single node.
- Training a Large Model when searching over many hyper-parameters
Scaling From Single Node To Apache Spark Cluster Computing
The following code block trains an SVM classifier on a mock classification dataset with 20 features(independent variables) in X & 1 labeled feature(dependent variable) in y. The GridSearchCV searches for the parameters by testing various SVM models. In this particular case, the param grid enables the search of 48 different model variants with different parameters to suggest the best model using the k-fold cross-validation technique. K-fold cross-validation involves randomly dividing the set of observations into k groups, or folds, of approximately equal size. The first fold is treated as a validation set, and the method is fit on the remaining k − 1 folds By default Grid Search in scikit-learn uses 3-fold cross-validation.
from sklearn.datasets import make_classification
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
import pandas as pd
X, y = make_classification(n_samples=1000, random_state=0
X[:5]
param_grid = {"C": [0.001, 0.01, 0.1, 0.5, 1.0, 2.0, 5.0, 10.0],
"kernel": ['rbf', 'poly', 'sigmoid'],
"shrinking": [True, False]}
grid_search = GridSearchCV(SVC(gamma='auto', random_state=0, probability=True),
param_grid=param_grid,
return_train_score=False,
iid=True,
cv=3
n_jobs=-1)
Grid Search in SciKit-Learn like any other API using the n_jobs parameter specifies how many concurrent processes or threads should be used for routines that are parallelized with joblib. When the n_jobs parameter is set to -1 like above in GridSearchCV, all CPUs are used. Additional details on how n_jobs drives single-node parallelism using joblib can be found at https://scikit-learn.org/stable/glossary.html#term-n-jobs. When we call the fit method to trigger the hyperparameter search below, parallelism is realized using all CPUs in a single node.
grid_search.fit(X, y)
When joblib-spark is used with scikit-learn, the grid search can scale to the distributed spark cluster and multiple models can be evaluated on multiple nodes to perform the hyperparameter search and parallel tuning. The following code block demonstrates how this parallelism can be achieved with minimal code change:
from sklearn.utils import parallel_backend
from joblibspark import register_spark
register_spark() # register spark backendΩ
with parallel_backend('spark',n_jobs=3):
grid_search.fit(X, y)Apache Spark ML
A common ML model Inferencing scenario is having a small or medium amount of labeled data to estimate a model from (e.g., 10,000 records), but a much larger unlabeled dataset to make predictions about.
In this scenario, one can train a model on a desktop/single server with a SciKit-Learn for ease of use and flexibility. Once trained, the model can be applied to the large unlabeled dataset by leveraging distributed computing with PySpark.
Scaling Scikit-Learn Model Inferencing Using UDFs
Using PySpark for distributed prediction is also a great fit if the ETL steps leading to inferencing are already implemented with PySpark.
With this in mind, we will take the curated model in Recipe 1 and explore how we can scale the inferencing using Apache Spark. For demonstration purposes, the code block below limits the number of mocked records in the spark data frame to 100, but it can scale to many millions of unlabeled data records that need to be inferenced/scored
import pyspark.sql.functions as F
from pyspark.sql.types import DoubleType, StringType, ArrayType
X_score, y_score = make_classification(n_samples=100, random_state=0)
best_model_from_gridsearch=grid_search.best_estimator_
n_features=20
column_names = [f'feature{i}' for i in range(n_features)]
pdf_X_score = pd.DataFrame(X_score, columns = column_names).reset_index().rename(columns = {'index': 'id'})
#Mocked Spark Dataframe with 100 mocked records for inferencing.
#In production this record could scale to millions
#and millions of records.
df_X_score = spark.createDataFrame(pdf_X_score)
@F.udf(returnType=DoubleType())
def predict_udf(*cols):
# cols will be a tuple of floats here.
return float(best_model_from_gridsearch.predict((cols,)))
df_pred = df_X_score.select(
F.col('id'),
predict_udf(*column_names).alias('prediction')
)
display(df_pred)Spark ML And Python Multiprocessing
Scikit-Learn with joblib-spark is a match made in heaven. As seen in Recipe 1, one can scale Hyperparameter Tuning with a joblib-spark parallel processing backend.
Spark itself provides a framework – Spark ML that leverages Spark’s framework to scale Model Training and Hyperparameter Tuning.
Hyperparameter Tuning
Studying Spark’s take on Hyperparameter Tuning gives great insights on using Python’s Multiprocessing library to scale the tuning process. It also gives an opportunity to understand how to extend this framework for cases where there are no out-of-the-box tools available – for example, Time Series Cross-Validation and hence, Hyperparameter tuning on Time Series Forecasting.
Performing k-fold cross-validation on Spark ML is straightforward.
Here is an example code snippet:
lr = LogisticRegression() grid = ParamGridBuilder().addGrid(lr.maxIter, [0, 1]).build() eval = BinaryClassificationEvaluator()cv = CrossValidator(estimator=lr, estimatorParamMaps=grid, evaluator=eval)
Here, the number of models that will be trained will be len(grid).
Upon peeking under the hood here, one can see the similarities between the JobLib-Spark model and Spark ML’s approach to hyperparameter tuning. See the appendix for condensed pseudocode.
In essence, for a k-fold cross-validation hyperparameter tuning run:
- At every iteration, all hyperparam combinations are evaluated in parallel
- Python Multiprocessing is used to submit parallel evaluations
- An evaluation is a Job in the Spark Session. The training and validation themselves use Spark for data-parallel training.
- The hyperparameters with the best metrics across iterations are returned.
Note that the parallelism will always be minimum(desired_parallelism, len(grid))
This knowledge comes in handy when tuning the learning program to fully utilize the cluster.
Hyperparameter Tuning with Time Series Cross-Validation
There are times when standard APIs cannot cater to the problem at hand. In such situations, if we know the underlying framework, we can extend it to the problem at hand.
For example, Time Series Cross-Validation.
Suppose we are tasked with training a model to forecast supply requirements at a warehouse. We will have to consider seasonality patterns, holiday patterns, etc to get an accurate forecast. This will require that the model be trained to understand the chronological features of the data.
As seen in the pseudocode from the appendix, Spark ML’s implementation of cross-validation assigns rows to folds at random. Randomness is encouraged in cross-validation. However, we cannot use the same for Time Series data. If the training instances(and folds) are out of chronological sequence, the algorithm won’t learn the model correctly.
Hence we need to extend the existing Hyperparameter tuning code to our needs.
There are many strategies for implementing a time series cross-validation. We want to capture seasonality patterns, hence training folds should not forget past data. This equates to the bottom-left quadrant in the diagram.
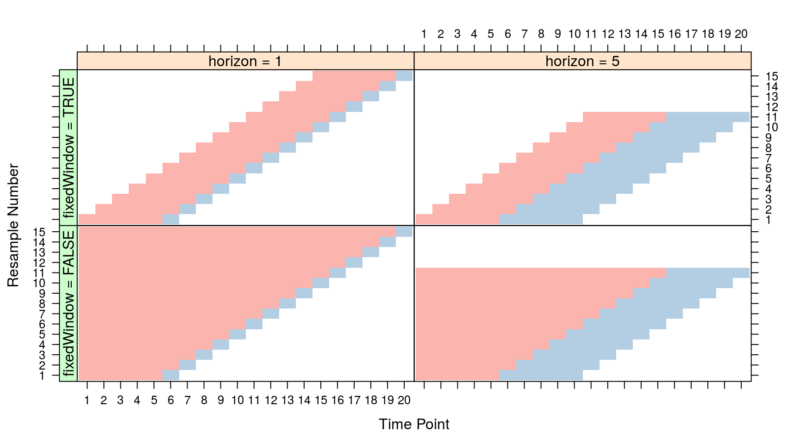
Rethinking the concept of “folds” of the cross-validation:
- Select the initial training fold (TS – TE). The validation fold(horizon) is (TE+1)
- Subsequently, the training folds increase by 1 Time Point. The validation folds move forward by a one-time point.
- This goes on until the end of the training data is achieved.
We can now override the “fit” method of the Spark CrossValidator to get a TimeSeries CrossValidator:
def _fit(self, dataset):
numModels = len(epm)
#Hiding redundant boilerplate code
#timeSeriesFolds is an array of date ranges. with each fold:
#The start of the training fold remains the same.
#The end of the training fold extends by one Time Unit.
#The validation fold moves forward by one Time Unit.
timeSeriesFolds = buildTimeSeriesFolds(dataset)
#dateCol=('date', 'start_date', 'end_date')
dateCol = self.getOrDefault(self.dateCol)
nFolds = len(timesliceData)
#Number of Time Units
horizon = self.getOrDefault(self.horizon)
#Indicate Desired parallelism to Python Multiprocessing
pool = ThreadPool(processes=min(self.getParallelism(), numModels))
for i in range(nFolds):
currentFold = timeSeriesFolds[i]
fold_start = currentFold[dateCol[1]]
fold_end = currentFold[dateCol[2]]
#Get folds from start date to end date
fold_data = dataset.filter((F.col(dateCol[0]) >= fold_start) &
(F.col(dateCol[0]) <= fold_end))
#Based on horizon size(Time Unit) get the date that will divide the training data from validation data
cutoff_date = self._determine_cutoff_date(fold_data, dateCol[0], horizon)
training_data, validation_data = self._timeseries_split(sliced_data, dateCol[0], cutoff_date)
training_data = training_data.cache()
validation_data = validation_data.cache()
tasks = _parallelFitTasks(est, training_data, eva, test_data, epm, True)
for j, metric, subModel in pool.imap_unordered(lambda f: f(), tasks):
#Rest of the code remains the sameTaking it to the next level…
This implementation of the Time Series Cross Validator still has an iterative component. In most scenarios, the number of folds is relatively low(SciKit-Learn defaults to 3) and hence this iteration is computationally insignificant.
However, in time series, the number of folds will increase linearly with:
- The increase in the date range of the training data.
- The increase in the granularity of the chosen time unit.
In this case, the iterative component of processing k-folds is going to slow down the Grid Search considerably. If the Parameter Grid itself is small, then the Spark cluster will be underutilized as not enough evaluations will be run in parallel.
Using what we learned about how Spark uses Python Multiprocessing, We can replace the iterative call
for i in range(nFolds):
With a multiprocessing pool as well. Since each evaluation is an independent spark job, we can parallelize this at will and use the Spark cluster to the maximum.
Implementing this can be taken up by the reader as an exercise!
Why Qubole
Scaling and speeding up Data Science workloads originally written in Python-based single-node compute frameworks like scikit-learn, doesn’t have to be very disruptive.
Like illustrated in Recipe 1 and Recipe 2 above, we can easily combine the strengths of frameworks like SciKit-Learn and Spark’s ability to scale computations across a distributed cluster to provide compelling solutions that boost performance and accelerate time to value.
Beyond this, even in the PySpark ML library, there are several opportunities to customize the evaluators and cross-validators to increase parallelism and gain much-needed efficiencies.
Qubole provides an Open Data Lake Platform that manages all of the infrastructure necessary to quickly develop data science pipelines, train, and deploy models leveraging Python and Pyspark. Signup for a Qubole free trial today.
Appendix
Pseudocode: Spark’s K-fold Validation Algorithm for Hyperparameter Tuning
from multiprocessing import Pool
#add a random column which assigns a row to a random fold
dataset := dataset.with_column(random(1..k_fold_count))
#in k_fold CV, we iterate over the training dataset k times
#in each iteration, 1 fold is selected as the validation set
#the rest of k-1 folds are used as the training sets
for k in k_fold_count:
validate_df := get_folds(k) #1 fold(kth)
training_df := get_folds(!k) #k-1 folds
#Generate learning tasks for m x n hyperparameter combinations
#Each learning task is a Spark Job
#Each task first "fits" the data in the training folds
#Then it "validates" the "fitted" model against the validation fold
parallel_learning_tasks := {
for combination in m_n_hyperparameters:
model = fit_data(training_df, combination)
metric = validate(validate_df, model)
}
#Python Multiprocessing Library initiates learning tasks in parallel
#The parallelism is decided based on the desired parallelism
with Pool(min(parallelism, len(m_n_hyperparameters)) as p:
metrics = p.map(parallel_learning_tasks)
#Save the best hyperparameter combination per iteration
save_best(hyperparam_combination)
#return the best performing hyperparameter combination
return_best(hyperparam_combination)


